U.S. scientists have applied a new technique to comprehensively analyze the human oral microbiome, which provides greater knowledge of the diversity of the bacteria in the mouth. For the first-time, scientists can provide high-resolution bacterial classification at the subspecies level, according to a new study in the Proceedings of the National Academy of Sciences (June 23, 2014).
The study will enable researchers to more closely examine the role of bacterial communities in health and disease.
 Gary Borisy, PhD.
Gary Borisy, PhD.Gary Borisy, PhD, a senior research investigator in the department of microbiology at the Forsyth Institute in Cambridge, MA, collaborated with A. Murat Eren, PhD, and Jessica L. Mark Welch, PhD, at the Marine Biological Laboratory in Woods Hole, MA, and also Susan Huse, PhD, at Brown University in Providence, RI.
The human body, including the mouth, is home to a wide range of microbial organisms. In fact, there are 10 times more bacterial cells in the human body than human cells, the researchers noted. Bacteria in nature live in complex, multispecies communities in which bacterial cells in close proximity to each other can exchange metabolic products and signals.
"Knowing who is living there and who their neighbors are is essential to understanding how they work together," Borisy stated in a press release. "We now have the opportunity to truly classify the bacteria and know what the distinct species are within these communities."
Study overview
The Human Microbiome Project, an effort of the National Institutes of Health, produced a census of bacterial populations from 18 body sites in more than 200 healthy individuals. DNA in these samples was sequenced from the gene in bacteria that encodes ribosomal RNA, called the 16S rRNA gene, or 16S. The 16S gene serves as a "barcode" for the identity of the organism.
“We now have the opportunity to truly classify the bacteria and know what the distinct species are within these communities.”
Although the project was groundbreaking in scope, interpreting the enormous numbers of barcodes obtained and distinguishing real barcodes from errors has been an issue. While general practice has been to collect similar barcodes together and put them into bins, this risks grouping together species that shouldn't be, such as pathogens and nonpathogens.
"The Post Office knows my name and address," Borisy said, "but imagine how it would respond to receiving letters (without addresses) for Borisi, Barisy, Borisey, or Borisky. Are these misspellings or real differences? That's a big problem facing the barcoders."
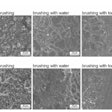
A new high-resolution method termed oligotyping overcomes this problem by evaluating individual positions in the barcode using Shannon entropy to identify the most information-rich nucleotide positions, which then define oligotypes. The team has applied this method to comprehensively analyze the oral microbiome.
Using the project's 16S rRNA gene sequence data for the nine sites in the oral cavity, the team identified more than 300 oligotypes. They associated these oligotypes with species-level taxon names by comparison with the Human Oral Microbiome Database. The scientists discovered closely related oligotypes, differing sometimes by as little as a single nucleotide that showed dramatically different distributions among oral sites and among individuals.
Many oligotypes were site-specialists, the researchers found. Some were preferentially located in plaque, others in gums or the cheek, and some were characteristic of throat, tonsils, tongue, palate, and saliva. The differing habitat distributions of closely related oligotypes reveal a level of ecological and functional biodiversity not previously recognized, they noted.
The Shannon entropy approach of oligotyping has the capacity to analyze entire microbiomes, discriminate between closely related but distinct taxa, and, in combination with habitat analysis, provide deep insight into the microbial communities in health and disease, the researchers concluded.